Connectase: An enzyme that fuses proteins in a specific manner
Proteins are central to almost all biological processes taking place within living organisms. However, proteins cannot last forever; they need to be regularly broken down and replaced. Controlled protein turnover is an essential activity for all living cells. A key facilitator within this pathway are proteasomes, large enzymes that degrade unneeded or damaged proteins through proteolysis, a chemical reaction that breaks peptide bonds.
Proteasomes are near universal. They are found inside all eukaryotes (higher organisms, such as protists, fungi, plants and animals) and archaea (single-celled organisms often found in extreme environments), and in some bacteria too. Due to their huge physiological relevance, Dr Adrian Fuchs and his team at the Max Planck Institute for Developmental Biology became interested in understanding more about their function and evolution. It was through this research that they discovered Connectase, a new protein with fascinating biotechnological potential.
How was Connectase discovered?
Dr Fuchs and his team were carrying out a bioinformatic analysis (a type of analysis used for understanding large and complex sets of biological data) when they discovered a protein that is very divergent from the proteasome, but still distantly related to it: Connectase. Curiously, this protein could only be found within methanogenic archaea. These microorganisms obtain their energy from producing large quantities of methane, a greenhouse gas, and are therefore key factors affecting our planet’s climate.
The researchers had expected Connectase to be a multi-subunit protease like the proteasome. However, their structural analysis showed that Connectase subunits do not assemble into proteasome-type complexes but rather exist as single protein chains, so-called monomers. Even more surprising, these monomers did not exhibit any proteolytic activity. Unlike other members of the proteasome family, Connectase showed only ligase activity. A ligase is an enzyme that can catalyse the joining of two large molecules by forming a new chemical bond. That essentially means it connects proteins together, rather than breaking them apart.
The researchers were able to show that Connectase specifically interacts with Methyltransferase A and is able to bind and
modify it.
How does Connectase function?
During further experiments with methanoarchaeal cell extract, the researchers identified a physiological substrate of Connectase in methyltransferase A (MtrA). MtrA is a key enzyme in methanogenesis, the pathway by which archaea produce methane. This pathway is thought to be one of the oldest processes in living organisms for energy generation and carbon fixation and an important contributor to the greenhouse effect.
The researchers were able to show that Connectase specifically interacts with MtrA and is able to bind and modify it. Further studies showed that only a short sequence in MtrA, the Connectase recognition motif, is required for this interaction. In fact, when this recognition motif was incorporated in a variety of other unrelated proteins, Connectase interacted with them just like with MtrA. The team was surprised to find that in both cases, Connectase does not only bind but also cleave this sequence into two fragments.
However, the reaction remained incomplete, no matter how the researchers altered the reaction conditions. This led them to the conclusion that the reaction must be reversible. It appeared that the recognition sequence was being constantly cleaved and reassembled.
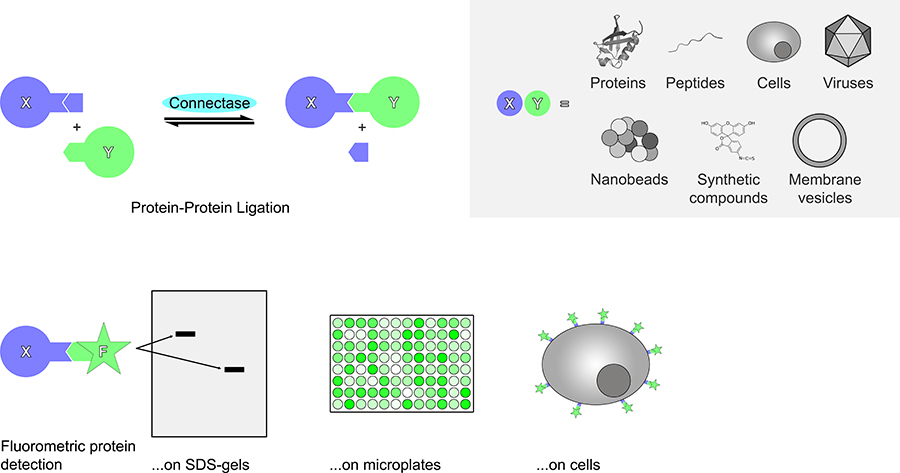
In order to prove this theory, Dr Fuchs and his team fused the recognition sequence to the N-terminal end of one protein and to the C-terminal end of another protein. Indeed, upon addition of Connectase, both proteins were fused together, with the recognition sequence bridging their N- and C-termini. This experiment showed that Connectase can be used to ligate any two proteins bearing the recognition sequence.
What are the biological implications of this discovery?
The unprecedented activity of Connectase was exciting for Dr Fuchs and his team, as it showed how versatile proteasome-like proteins could be. It also helped them to understand the link between their diverse reactions: proteasome subunits are made as precursors that must be activated (“autolysis”) before they can degrade proteins (“proteolysis”) or shuffle peptides (“splicing”). All these reactions are mechanistically related and the Connectase reaction is a fourth, unprecedented variation of this reaction scheme that sheds a new light on the different proteasome functions.

It is yet enigmatic how exactly the ligase activity of Connectase is affecting methane production. For this process, it is essential that the Connectase target MtrA is located at the membrane, a lipid layer that encapsulates the cell. The Connectase recognition sequence is located between the functional core of MtrA and its membrane anchor. Consequently, the Connectase reaction may control or alter methane production, by separating and re-ligating the two MtrA fragments.
While these exciting prospects are subject to ongoing studies, the capability of Connectase to ligate proteins makes it highly attractive for a variety of biotechnological applications.
What advantages does Connectase have over other enzymes?
There are several existing methods that we can use to ligate proteins, but they all come with different advantages and limitations and are therefore useful in different settings. Connectase-mediated ligations are not quite as traceless as some of these other ligation methods. They necessitate a ~20 amino acid recognition sequence, so Connectase will only be used in cases where the presence of this sequence can be tolerated. In these cases, however, Connectase offers a combination of simplicity and efficiency that is hard to rival.
Compared to other established protein ligases, Connectase is far more specific, meaning that it only ligates proteins that bear the recognition sequence. It does not act on other proteins “by mistake”. It is therefore highly active even in very dilute and impure solutions. While side-reactions are a major issue when using other protein ligases, Connectase uses a different reaction mechanism that prevents such unintended functions.
Connectase is also easy to handle: reactions don’t need lengthy optimisation procedures, complicated set-ups, special equipment or defined buffer systems. Substrates can be produced recombinantly but are also amenable to chemical synthesis. This makes Connectase a valuable tool for developing new methods.
Connectase’s unique characteristics make it highly attractive for a variety of biotechnological applications.
What biotechnological applications could Connectase have?
The most obvious use of Connectase would be to fuse different types of protein together to create entirely new ones. This could lead to hybrid proteins that would have been difficult to produce by conventional means, such as new antibody conjugates with a diverse set of binding sites.
Connectase can also be used to control the location of proteins. For example, proteins can be attached to the cell surface, on microbeads, on virus-like particles or on surfaces for cryo-electron microscopy in a geometrically defined way.
Finally, Connectase can introduce chemically synthesised molecules into proteins. In pharmacology, this is of use when creating antibody-drug conjugates, which can be used to bring drugs specifically to cancer cells. Alternatively, proteins can be labeled with fluorophores. This would allow for their detection intracellularly or on the cell surface. It would also help for proteins to be detected in SDS-gels, on microplates, and in affinity measurements.
Conclusion
In summary, Connectase represents a fascinating scientific find. From a purely theoretical perspective, it’s interesting to study its unique characteristics and differences from other proteasome structures. Still, most exciting is the potential for Connectase’s unique characteristics to be used in a range of future biotechnological applications.
Personal Response
In which area of biotechnology do you believe Connectase could have the most exciting impact?
<> We think that many academic scientists could find the system useful to tackle their research questions, but it may also be employed in industrial applications. Specifically, we are currently using it to detect proteins in complex solutions or on living cells. While this is usually done with fluorophore-labeled antibodies, Connectase can directly ligate such fluorophores to a protein of interest. In preliminary assays, we find that the Connectase-based method allows a more rapid, sensitive and specific detection of proteins.