Integrating molecular epidemiology and microbiome science
How cancer patients respond to therapy
Despite numerous advances in treatment and continuing efforts by researchers across the world, cancer continues to destroy lives. Treatment regimens can often be taxing and unfortunately, some patients do not respond well to the medications on offer and may develop adverse side effects as a result of treatment.
This unacceptable reality has led researchers to seek a way to predict whether a patient will respond to a cancer treatment before it is administered. The answer to this huge question may lie with our microbiota: the cumulative collection of microbial organisms on and in the body. Recent research has shown that there is a link between receptiveness to cancer treatment and microbial composition. The developments in this field have even led to the suggestion that the microbiome may be able to be manipulated to optimise the effectiveness of cancer therapeutics. However, there is much to be done to further our understanding of the microbiome in cancer and its potential clinical utility. The biological mechanisms underlying the connection between the microbiota and treatment response remain understudied, and there continues to be a lack of consensus in which microbes drive differences between treatment responders and non-responders.
The team behind the ideas
Within the field of microbiome science many researchers are exploring different mechanisms by which the human microbiome and cancer cells interact. At the Moffitt Cancer Center in Tampa, Florida, USA, one of the lead researchers in this field is Dr Christine Pierce (Assistant Member in Cancer Epidemiology). Dr Pierce has worked within the field for many years, gaining her PhD from the University of Michigan and completing a post-doctoral fellowship at Moffitt Cancer Center. Her research uses integrative molecular epidemiological techniques to understand the human microbiome and cancer.
In her latest research she was ably assisted by an excellent research team including Shirlene Paul, research data analyst, and Stephanie Hogue, research coordinator. Other key collaborators at Moffitt Cancer Center include Dr Lary Robinson, Dr Jhanelle Gray, Dr Jimmy Caudell, Dr Jose Conejo-Garcia, Dr Youngchul Kim, Dr Stephanie Schmit, and Dr Anna Giuliano, as well as other colleagues at the University of Florida (Dr Christian Jobin and Dr Raad Gharaibeh) and the Jackson Laboratory for Genomic Medicine (Dr George Weinstock, Dr Erica Sodergren, and Dr Bo-Young Hong).
Within the field of microbiome science many researchers are exploring different mechanisms by which the human microbiome and cancer cells interact.
Bridging fields in microbiome research
At the Moffitt Cancer Center, Dr Pierce’s research is largely focused on incorporating microbiological, epidemiological, and immunological principles to uncover the associations between the local tumour and oral microbiomes, the systemic gut microbiome, and patient responses to cancer therapies. Several studies on different cancer types have been conducted to support these aims.

In studies on patients with head and neck cancer, the interaction between the local tumour microbiota and treatment related complications were examined. Head and neck cancer patients are likely to develop debilitating side effects during concurrent chemotherapy and radiation treatment. Unfortunately, this method of treatment, known as chemoradiation, is the most effective treatment for head and neck cancer sufferers who are typically diagnosed in the later stages of the disease.
These side effects include oral mucositis – large, painful ulcers throughout the oral cavity, and oral candidiasis – a fungal overgrowth of the typically non-harmful Candida albicans species. Frozen, archived tissue samples collected from patients prior to treatment were evaluated for microbial composition and tumour infiltrating lymphocyte levels to elucidate relationships between the microbiome, anti-cancer immunity, and treatment outcomes. A thorough review of patient medical records was conducted to determine treatment response and development of treatment related side effects. The demographic and health history of patients was also recorded to contribute more data to this pilot study.
The team found that the fresh frozen head and neck cancer tissue samples were a suitable source of data for future research projects and identified this source as a potential method for predicting which patients would experience adverse treatment outcomes.
In another project, the Moffitt Cancer Center researchers are studying the oral microbiome of oropharyngeal cancer patients to determine if there is an association between oral microbiota and clinical response to chemotherapy and radiation. Oropharyngeal cancer prevalence is on the increase in the United States with five-year survival rates remaining at 30-60%. The treatment for oropharyngeal cancer is similar to that of other head and neck cancers and can result in similarly debilitating incidences of mucositis and candidiasis.
…researchers now have more data sources available to explore the impact of cancer treatments on microbiota, and vice versa.
Oral gargles from 60 recently diagnosed patients were obtained prior to treatment and examined to identify the levels of predictive microbiota. Data revealed that patients with highly diverse oral microbiomes prior to treatment had an increased likelihood of developing mucositis but a decreased likelihood of developing candidiasis. Further research evaluating the functionality of oral microbes and the influence of oral microbiota on immune and inflammatory responses may help to unravel the complex relationship between the microbiome and cancer treatment outcomes.
Collecting patient data at home
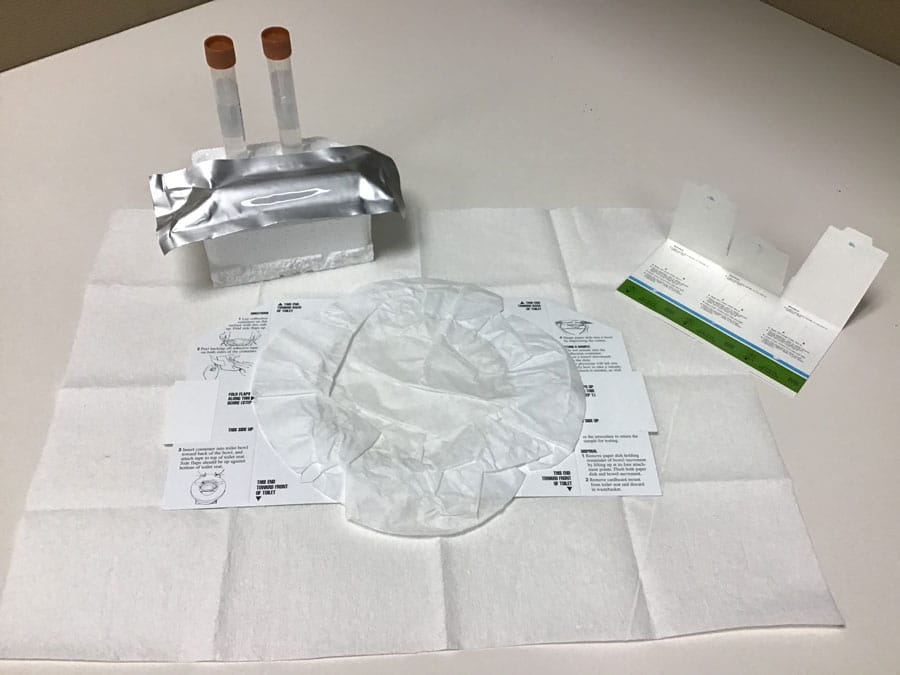
The team have developed an at-home method of stool collection to study the relationship between the gut microbiome and response to cancer immunotherapies. This project received support from The Florida Academic Cancer Center Alliance and Hoenle Foundation who have largely funded the development of the stool collection kit and the corresponding pilot study to gather patient data. The H. Lee Moffitt Cancer Center and Research Institute also contributed shared resources to this research.

The development of an at-home method for stool collection has several advantages. Primarily, the kit is easy for patients to use and allows a few days for specimen collection, ensuring patients with constipation have time to collect a sample. Room temperature storage eliminates the need for cumbersome ice packs or storage in a household freezer. Additionally, several downstream analytical platforms are compatible with the kit components, enabling a holistic view of the gut microbiome to be obtained. The kit is also easily assembled and inexpensive, making it ideal for large population studies.
The kit, assembled with aseptic practices in mind, contains three different collection platforms. The first is a faecal occult blood test (FOBT) card. This card is comprised of three slides, each containing two filter paper windows where sample is applied. The FOBT card has previously been validated as an appropriate method for use in microbial metagenomics to determine microbial composition in stool samples. The second and third collection methods are small 15ml faecal collection tubes. The first tube is filled with 8ml of 95% ethanol, a medium particularly recommended for faecal metabolomics studies. The second tube contains 8ml of RNAlater, a preservative used to stabilise RNA for up to six days without freezing. It is the most widely recommended preservative for metatranscriptomics studies.
Out of 64 late-stage lung cancer patients approached over an eight month period from January to August 2018, 53 were willing to contribute samples for research. Over half of the consented participants (31/53) were able to provide a sample prior to treatment start. Preliminary analyses on a sub-sampling of patients showed that Ruminococcus was significantly associated with clinical response to immune checkpoint inhibitor therapy. This genus also down regulated genes associated with energy production and conversion. The combination of metagenomics, metatranscriptomics, and metabolomics analyses will provide a holistic image of the gut microbiome’s structure and function in relation to response to immunotherapy in lung cancer.

Future directions
Being able to better understand the specific characteristics of multiple cancers means that researchers now have more data sources available to explore the impact of cancer treatments on microbiota, and vice versa. Going forward, the Moffitt Cancer Center team hopes their unique and well-considered collection kit will encourage more population scientists to value the importance of adding standardised stool sample collection to their research.
Certainly, cancer is not disappearing overnight and any research which can help to make the treatment of such devastating illnesses more successful should warrant further investment.
Personal Response
Has developing this kit led you to consider any other methods of data collection, particularly those of patient samples, which could be improved?
<> Yes! In our efforts to expand upon the current collection kit, we identified a preservation method that maintains bacterial viability at room temperature. Now, our stool collection kit is able to preserve one stool sample in four different types of media for four different analytical purposes. Live bacteria collected from the guts of cancer patients can be used in experiments with animal models, which will allow us to investigate how certain alterations to the gut microbiota can change cancer treatment outcomes.