Using metabolomics and artificial intelligence to explore the universe of the dark metabolome
Metabolism is a term used to describe chemical reactions in the body, and the comprehensive, high-throughput study of metabolism is referred to as metabolomics. There are two main types of metabolism; catabolism is the breakdown of molecules to obtain energy, and anabolism is the synthesis of compounds which are required by cells of the body. In addition, metabolism can refer to the activation, detoxification and elimination of foreign or unwanted substances.
In humans, metabolism is a complex process involving thousands of different enzymes which break down different foods, drugs and contaminants into smaller chemical compounds which can be used, excreted or further transformed to other substances by specific enzymes in the body (biotransformation). These biotransformation enzymes are produced by different organs in the body, including the liver, kidney, stomach and intestine, as well as by bacteria which live in the intestine (the microbiome).
The end result of this enzyme-driven process is hundreds of thousands of different chemicals (metabolites) circulating around the body. Metabolites can be beneficial or detrimental to the body; some metabolites provide energy for cells, whereas others are toxic and must be removed from the body, for example viaurine. Professor David Wishart at the University of Alberta has been involved in the discovery and cataloguing of thousands of these metabolites for the past 15 years.
The dark metabolome
Despite their importance in the day-to-day functioning of the human body, scientists can only identify about 2% of metabolites at present, with the remaining 98% being referred to as the ‘dark matter’ of the metabolism, or the ‘dark metabolome’. Although it was previously thought that scientists had a thorough understanding of metabolic processes, improvements in analytical chemistry technologies have led to the detection of many new, unknown metabolites. These unknown compounds are frequently detected using today’s scientific instruments but cannot readily be identified and therefore limit scientists’ current knowledge of human metabolism. It is this dark metabolome that Professor Wishart seeks to shine some light upon. Identifying these unknown metabolites is expected to increase our understanding of human physiology, disease and biochemistry. Furthermore, including the soil and aquatic microbiome in tracking these metabolic biotransformations will shed light on the complex interactions and the metabolic ‘life cycle’ between humans and the environment, for example the impact of pesticides on human health.
Identifying previously unknown metabolites will increase understanding of human physiology, disease and biochemistry.
Although much effort has gone into investigating the metabolic processes involved in the production and breakdown of essential metabolites, such as fats and proteins, less work has focused on non-essential metabolites. These are the chemicals that are thought to make up much of the dark metabolome, and which are more often not derived from catabolism or anabolism, but from the biotransformation of other substances called xenobiotics. Xenobiotics are foreign substances such as drugs, plant chemicals, food additives or pesticides. There are hundreds of thousands of xenobiotics – and potentially millions of xenobiotic metabolites.

Using machines to replace man
One way that unknown metabolites can be identified is through the use of computational techniques, such as artificial intelligence and machine learning, to predict the metabolism, or biotransformation of chemicals in the body (in silico metabolism). Although it may sound like something out of a sci-fi film, machine learning is the term used to describe the use of artificial intelligence to enable systems to automatically learn and expand on previous experiences without being specifically programmed, using algorithms and statistical models, rather than human instruction to guide completion of tasks. The benefits of excluding human intervention are that systems can analyse bigger, more complex data faster and more accurately than would otherwise be possible. Isolating, purifying and determining the structure of a single unknown metabolite can take a skilled chemist one to two years. Using an in silico approach, this process can take a matter of minutes. If the dark metabolome consists of one million compounds (as currently thought), scientists predict it will take hundreds of years to characterise it manually. A faster, computational approach based on metabolism prediction appears to be the only option.
Metabolism prediction programmes typically start with a parent molecule and employ pattern recognition techniques, hand-made rules, or machine learning methods to identify sites of metabolism on the parent molecule, the likely metabolic reactions, and the chemical products resulting from metabolism at that site. Although a number of tools for metabolism prediction have already been developed, these were mainly designed to aid pharmacology-focused studies. As a result, today’s metabolism prediction tools are not really suited for shedding light on the dark metabolome.
What is BioTransformer?
Professor Wishart and his team have used cutting-edge machine learning technology to develop a freely available software package that is specifically designed to help reveal the dark metabolome, called BioTransformer.

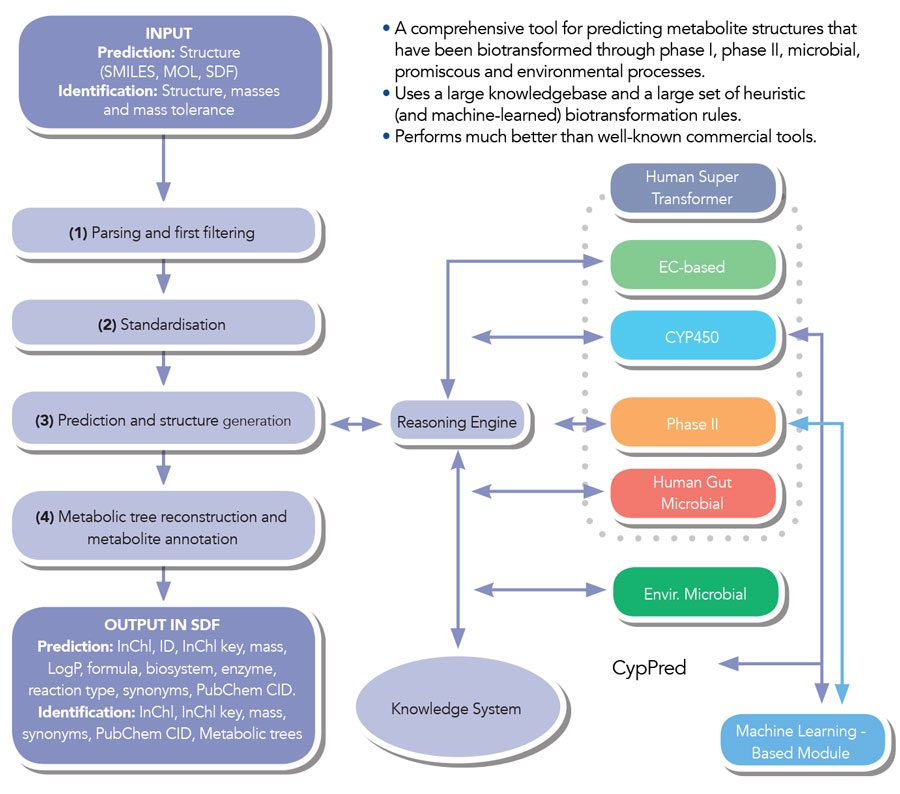
The function of BioTransformer is two-fold; to predict small molecule metabolism using a metabolism prediction tool (BMPT), and to characterise unknown metabolites using a metabolite identification tool (BMIT). BioTransformer predicts small molecule metabolism in mammals, as well as the gut microbiome, soil microbiome and aquatic microbiome. More relevant to mining the dark metabolome is the ability of BioTransformer to use metabolism prediction to assist scientists in the identification of unknown metabolites.
BioTransformer harnesses existing knowledge about known enzymatic reactions and uses pattern recognition and machine learning techniques to accurately predict the metabolism of almost any known chemical, including xenobiotics. The team at the University of Alberta developed and encoded almost 800 biotransformation rules, over 1,700 enzyme-based reaction rules, and multiple metabolic pathways in the BioTransformer knowledgebase.
Alongside the information retrieved from the knowledge base, a reasoning engine was incorporated to implement reaction site prediction and selection, as well as chemical product prediction. BioTransformer uses decision trees for its reasoning engine. Decision trees model decisions and their possible consequences, similar to the yes/no quizzes often found in magazines, and can select the most likely biotransformations or pathways for a particular situation.

BioTransformer consists of five independent reaction prediction modules which can be selected or combined together by the user to meet specific research queries: promiscuous enzyme-based, CYP450 (or Phase I), Phase II, human gut microbial, and environmental (soil or aquatic) microbial. Each of these modules refers to a different set of metabolic reactions, for example CYP450 enzymes principally work in the liver and are responsible for first pass (Phase I) metabolism of many substances, including drugs; while Phase II reactions further transform, or lead to excretion of metabolites resulting from initial CYP450 metabolism.
Professor Wishart describes BioTransformer as a ‘rocket ship’ to help biochemists explore the unknown chemical universe within the human body.
To be useful for analysing the dark metabolome, BioTransformer had to be very thoroughly tested and optimised to give vitally accurate metabolic predictions for a wide range of xenobiotics and endogenous metabolites. Indeed, Professor Wishart has shown that BioTransformer is capable of outperforming other commercially available tools, with results up to three times better than existing programmes.
Computational simulation of the process of human metabolism by BioTransformer allows scientists to rapidly predict hundreds of thousands of novel metabolites from the original chemicals. These computationally predicted, biologically feasible metabolites are now being used by Professor Wishart and his collaborators to begin the characterisation of the dark metabolome. In fact they have already used BioTransformer and mass spectrometry techniques to help identify a dozen previously identified xenobiotic (plant) metabolites in urine.

The rocket ship of metabolomics
Professor Wishart describes BioTransformer as a ‘rocket ship’ that enables biochemists to explore the unknown chemical universe within the human body. One major benefit to the software is that it is open access and freely available, meaning that expensive licensing arrangements do not present barriers to research. This will encourage more researchers to use the tool and will foster an online, idea-sharing community that can progress understanding of the dark metabolome.
Potential applications of advancing the capabilities of in silico metabolism prediction software include expansion of chemical databases of drugs, food compounds, environmental contaminants and other biologically and environmentally relevant chemicals. Indeed, BioTransformer has the potential to discover new biomarkers, bioactive metabolites and to inform development of drugs and household products, as well as facilitating the characterisation of the dark metabolome.
Personal Response
How much work does it take to keep BioTransformer up to date?
<>We are dedicating two full-time staff members to keep BioTransformer up to date and to continue to improve its accuracy, speed and performance. It’s proven to be much more popular and more useful than we expected.
As with other software, will upgrades be required in the future?
<> Certainly, as we learn more about metabolism and as we find chemicals in the dark metabolome that were completely unexpected or unpredicted, we will have to upgrade and improve BioTransformer. It will always be evolving – at least until we’ve solved the problem of the dark metabolome.
