Synthesising circular RNA is as easy as PIE
Naturally occurring circular loops of RNA, circRNAs, are the result of rare but significant backsplicing events. They are found throughout the eukaryotic domain and have been associated with pathological states such as tumour growth and suppression. Existing molecular techniques have taught us a great deal about circRNAs, but they are often difficult to implement and limited in scope. Dr Jason W Rausch of the National Cancer Institute at Frederick, Maryland, USA, has developed a new protocol for synthesising large, complex, artificial circRNAs with great precision, providing an invaluable tool for future research in the field.
Several decades ago, researchers first reported instances of ‘scrambled exons’, wherein the order of protein-encoding exon segments of some cellular RNA in higher organisms did not match the corresponding order of the amino acids in the proteins they were purported to encode. Since then, circular loops of RNA, or circRNAs, have been discovered in many different eukaryotic groups, from fungi to fish, birds to insects, and plants to protists. Originally, circRNA was thought to be merely a mistake in transcription; more recently, however, circRNAs have been shown to possess significant functionality in areas such as cell division and neurogenesis, and can also act as biomarkers for major diseases such as Alzheimer’s, diabetes, and neurological conditions.
Of the more than 100,000 human circRNAs that have been discovered to date, functionality has been attributed to only a handful, while the roles played by the vast remainder continue to be a mystery. This is partly because it is sometimes difficult to distinguish circRNAs from their linear RNA counterparts using existing methodologies, and because current approaches for producing synthetic circRNAs can be cumbersome, inefficient, and imprecise.

Making circRNA in nature and in the laboratory
CircRNAs are curious creations. They are produced naturally in cells by a process called ‘backsplicing’, which occurs when the cellular splicing machinery unusually connects the 3’ end of an exon (a coding segment of a gene) to the 5’ end of the same or a different upstream exon, creating a closed loop of seemingly misfit exonic RNA with no obvious purpose. Because circRNAs lack termini, they cannot be recycled by cellular enzymes that work by binding to an end of ‘linear’ RNA and chewing it back until there is nothing left. As a result, the cellular half-lives of circRNAs are much longer than their linear counterparts. Their closed structures also serve to stabilise circRNAs, likewise contributing to their long intracellular half-lives and making synthetic versions excellent building blocks for construction of geometric RNA nanoparticles – state of the art designer tools for delivery of small therapeutic RNAs to intracellular targets, safe vectors for genetic engineering, and even potential mimics of naturally occurring circRNAs with utility in studying their functions.
Synthesising circular RNA using permuted Group I introns had previously been limited by sequence restrictions.
Although there is great potential in each of these applications, producing synthetic circRNA in abundance and with high precision and purity has not been without its challenges. Conventional synthesis methods involve multiple components, are typically labour intensive, inefficient, and imprecise, and generate products difficult to purify from non-circular RNA precursors. Moreover, these difficulties expand exponentially as the circRNAs to be synthesised become larger and more complex.
Fortunately, many of these problems can be solved by exploiting the inherent activity of ribozymes – segments of RNA possessing enzymatic activity – called Group I introns. These remarkable RNA elements can excise themselves from within the longer pieces of RNA in which they are nested while simultaneously joining the flanking segments together. Moreover, they do so spontaneously and without the aid of proteins or other dedicated cellular components. By engineering these Group I introns so they are genetically ‘split’, with the two halves flanking an internal RNA segment (rather than itself being flanked), their end-joining function can be redirected to link the termini of the internal RNA segment to create a synthetic circRNA. Collectively, the RNA segment to be circularised, flanked by the two halves of the split Group I intron, is called a permuted intron-exon (PIE) construct. This device can be used to synthesise circRNA with great efficiency and precision in a test tube, and automatically, with only minimal intervention by the researcher required.
Development of the PIE technology facilitates synthesis of designer circRNA gene expression vectors, including some that have the potential to functionally resemble the highly successful mRNA COVID-19 vaccine vectors produced by Moderna and Pfizer. Unfortunately, two small but functionally important segments of the split Group I intron in PIE constructs overlap with the sequence of the internal RNA to be circularised, limiting design flexibility. Although this seemingly minor sequence requirement does not meaningfully affect circRNA gene synthesis constructs, it can significantly restrict researchers’ design options for most other applications. Consequently, the PIE approach must be thought of as having limited utility if it cannot be used to synthesise circRNAs containing the exact sequences a researcher requires.
Optimising the Permuted Intron-Exon approach
To maximise the potential utility of PIE constructs, Dr Jason W Rausch from Maryland’s National Cancer Institute at Frederick, USA, recently decided to both characterise these sequence restrictions and devise ways to overcome and circumvent them.
Part of Dr Rausch’s analysis involved systematically introducing changes into the E1 and E2 segments of a control PIE construct and measuring how these changes affected circRNA synthesis specificity and efficiency. His results were surprising. Of the combined ten nucleotides comprising the native E1 and E2 sequences, almost any nucleotide substitution was well tolerated for all but four, and limited substitution was acceptable for all but two. These findings were further supported by in vitro selection experiments in which the E1 and E2 sequences were randomised, and the variants capable of producing circRNA catalogued. These results likewise demonstrated that although there is much greater sequence flexibility in E1 and E2 than previously believed, Group I intron functionality did require some sequence specificity at two positions in these segments and near-absolute specificity at two others. Further investigation showed that even greater sequence flexibility could be achieved by changing other parts of the split intron sequence to compensate for mutations in E1, thereby maintaining the local RNA structure critical for intron function.
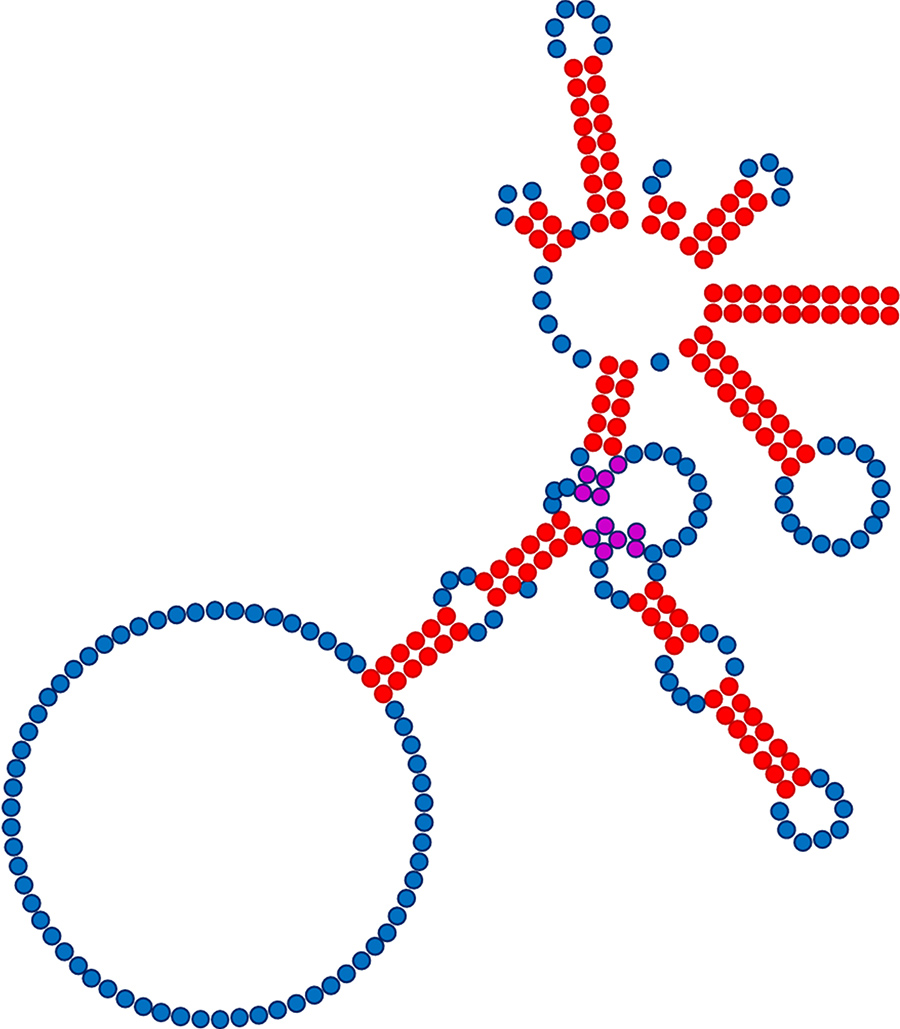
Taken together, Dr Rausch’s work showed that the number of restrictive nucleotides in the PIE construct was not ten but four, or even two, so long as compensatory changes designed to maintain Group I intron structure were also introduced. These findings both enhance our understanding of the sequence tolerances of the Group I intron analysed and greatly expand our capacity to customise PIE constructs for synthesis of circRNA having almost any sequence a researcher requires.
Dr Rausch also worked to develop a parallel approach in which the sequence of the RNA to be circularised is permuted to accommodate the E1–E2 sequence requirements of a PIE construct instead of the other way around. In theory, because circRNAs have no termini, it shouldn’t matter which nucleotides in a circRNA precursor sequence are joined by the activity of the split Group I intron, merely that the resultant circRNA contains all the required RNA sequence in the correct order.
Recent methodological advances now permit synthesis of exact replicas of human circular RNAs with unprecedented precision and scale.
Dr Rausch tested this theory using the sequence of circPVT1, a naturally occurring 410-nucleotide human circRNA shown to be involved in regulating cellular senescence and aging. Whereas natural circularisation of circPVT1 occurs by backsplicing, in which nucleotides 1 and 410 of a PVT1 gene exon are ligated together by the cellular splicing machinery, this exon sequence was reordered in the context of the PIE construct precursor (330–410,1–329) for end joining between nucleotides 329 and 330. This was done to exploit the nearly identical sequences of native PVT1 exon nucleotides 324–331 and the E1–E2 segments in the Group I intron, thus both preserving the functionality of the ribozyme and incorporating only the desired native sequence into the nascent, synthetic circPVT1 RNA. More generally, and prosaically, this re-engineering offers a means by which researchers can have their PIE and eat it too.
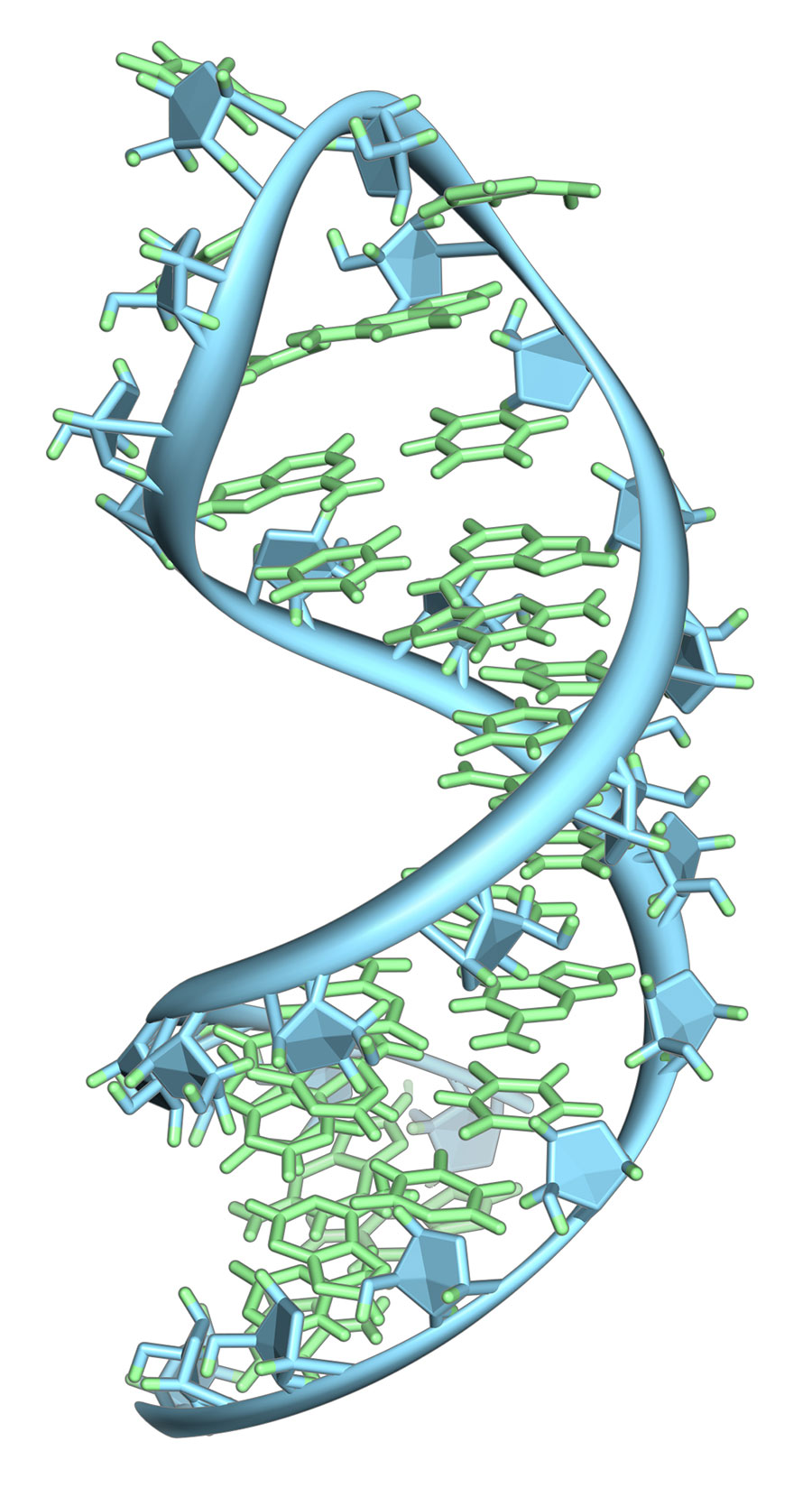
The experiment was an unqualified success. The circRNA yield was robust, and the synthetic circPVT1 sequence precisely matched the native, as verified by next-generation sequencing. The utility of the approach was then validated by exploring the structure of synthetic circPVT1 RNA by atomic force microscopy. An equivalent analysis of native or synthetic circPVT1 produced by conventional means would be virtually impossible, given the limitations of yield and purity restricting these methods.
The future of PIE
So, what can be done with synthetic circRNAs now that wasn’t possible before? As previously mentioned, the relative structural rigidity and durability of circRNAs, together with a now much broader range of variants that may be easily synthesised, greatly expands their potential utility as RNA nanoparticle building blocks, or even ‘dumbbell’ structures mimicking but more durable than siRNAs – short, naturally occurring, double-stranded RNAs that serve many regulatory functions in human cells. Additionally, the yields achievable by PIE-mediated circRNA synthesis opens the door for previously untenable structural analysis of even long, complex human circRNA variants by NMR, X-ray crystallography, cryo-EM, or, as demonstrated, atomic force microscopy.
Perhaps the most promising application of this more refined version of PIE is for transfection of perfect sequence mimics or mutant variants of important native circRNAs into living cells. For instance, one can imagine transfecting synthetic circ-Foxo3, found in breast cancer tissue and shown in some circumstances to inhibit tumour growth, into tumour cells in culture or animal models, then measuring the effects on cellular growth and spread. The therapeutic potential of cir-ITCH, a circRNA shown to inhibit progression of lung and oesophageal squamous cell carcinoma, might be similarly evaluated. Finally, the ease with which synthetic circRNA can now be genetically modified yet still produced in abundance permits facile analysis of mutant variants, both to determine which segments of a given circRNA are critical for function and even to improve function and create variants of greater potency. In short, these advances in PIE technology provide avenues for study and even therapeutic applications of human circRNAs that were previously inaccessible.

Personal Response
Some of these concepts might appear completely alien to those without a thorough knowledge of molecular biology – where do you take your inspiration from?
To advance science and improve the human condition, I enjoy both pushing the limits of existing technologies in molecular biology and helping to develop new ones. The future is bright for those scientists and clinicians who embrace technological advances, and I take satisfaction in playing even a small part in helping to pave the way for the next generation
