How to make a CRISPR cow
For centuries, breeders have selected animals displaying desirable qualities to be the parents of the next generation. When a breeder chooses an animal based on a visible characteristic – for example, coat colour – they are actually selecting between the naturally occurring variations in DNA sequences that are responsible for differences between individuals.
Using conventional breeding, achieving the desired changes can take decades. This is especially true of large livestock species like cattle, which have long generation intervals. Advances in genome editing offer an opportunity to bring about useful changes far more quickly. In this process, specific DNA sequence variations can be inserted directly into livestock genomes.
One common method used to edit genomes is the CRISPR/Cas9 system. This system has two components. The Cas9 protein, which cuts DNA, and a guide molecule that directs Cas9 where to cut. Bound together, they form a complex that cuts DNA at the target site. The resulting break, called a DNA double strand break (DSB), is then fixed by the cell’s natural DNA repair machinery.


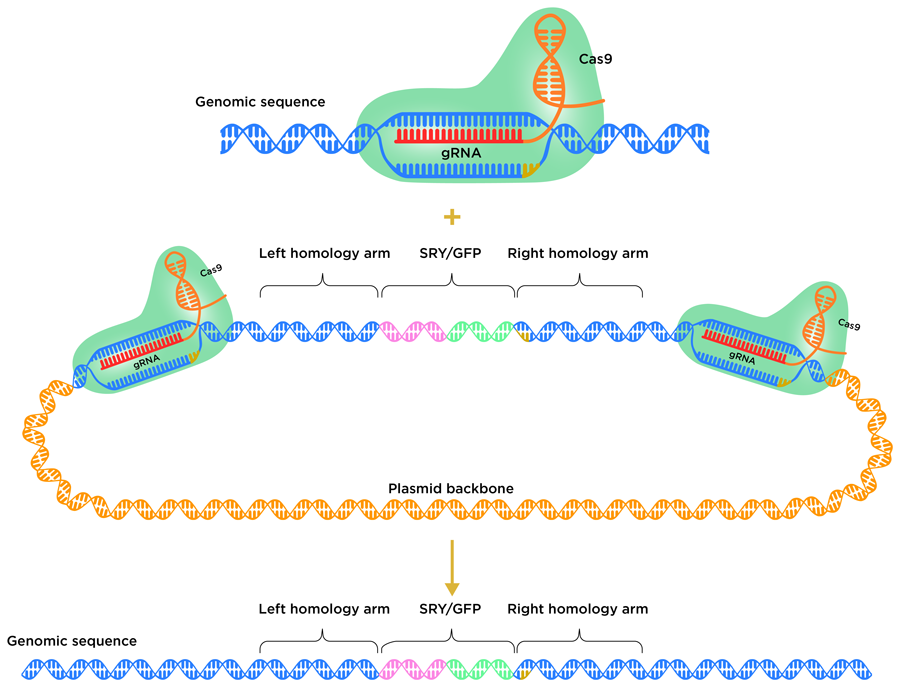
Cells use different natural mechanisms to repair DSBs. The nonhomologous end joining (NHEJ) pathway and the homology-directed repair (HDR) pathway are two possible repair routes. NHEJ typically leads to short insertion/deletion events (where a tiny fragment of DNA is inserted into or deleted from the genome) near the cutting site. Alternatively, HDR can be used to introduce useful DNA sequences from a donor template. The HDR donor repair template typically harbours the DNA fragment or gene to be inserted, flanked on either side by homology arms. These arms mirror the sequence of the DNA on either side of the CRISPR target site. So far, it has proven to be very difficult to insert whole genes into livestock genomes using the HDR pathway in embryos.
To avoid the insertion of the new gene being thwarted by the cell’s DNA repair system, the team employed a homology-mediated end joining (HMEJ) strategy.
Recently, a team led by Dr Alison Van Eenennaam and PhD student Joseph Owen at the University of California, Davis, used the CRISPR-Cas9 system in early bovine embryos to produce a calf carrying targeted insertions of the SRY gene. The bull calf, named Cosmo, was born in April 2020 during the COVID-19 pandemic. Cosmo is unique in that he also carries copies of a green fluorescent protein gene. Fluorescence was used as a marker to identify embryos carrying the targeted SRY gene insert. Those embryos were then transferred to surrogate cows, and one subsequently gave birth to Cosmo.
Why this gene?
The SRY gene carries the instructions for a protein called the sex-determining region Y, which is involved in male sexual development. Specifically, this protein acts as a transcription factor, which means it attaches to specific regions of DNA and helps control the activity of particular genes. The protein starts processes that cause a foetus to develop a male gonad (testes) and prevent the development of female reproductive structures (uterus and fallopian tubes). Inheriting the SRY gene causes an embryo to develop as a male. Bulls that could produce all-male offspring would be desirable for beef farmers; in the same way, bulls that sire all-female offspring would be desirable for dairy farmers.
Designing the guide
As a first step in the insertion of the SRY gene into the embryo, the team developed a guide RNA. This guide RNA (single-stranded nucleic acid, whereas DNA is double-stranded) directed the Cas9 protein to cut at a particular site on bovine Chromosome 17. This site, H11, is a known “safe harbour”, meaning that no essential genes are disrupted by gene insertions at this location. The team predicted that the guide would instruct Cas9 to cut at the target H11 site, and not at other locations, because it did not match any other bovine DNA sequences. This design helped to ensure that Cas9 would not cut at an “off target” location in the genome. The guide was tested on the target sequence both in vitro, and in vivo in a bovine cell line, to ensure it directed Cas9 to efficiently introduce DSB in the DNA at the H11 target loci.
Timing is everything
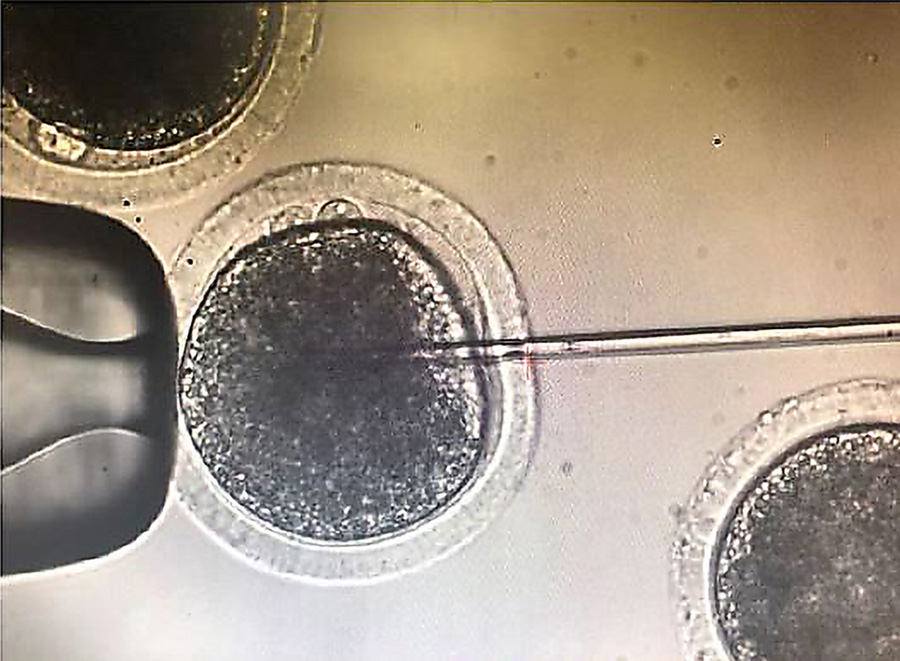
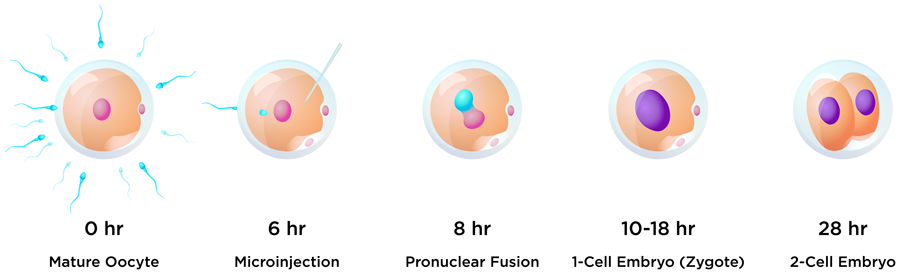
One of the aims of this research was to edit early embryos immediately after fertilisation to avoid a condition called genetic mosaicism. Mosaicism occurs when two or more groups of cells with different genotypes are present within an individual that has developed from a single fertilised egg. This usually happens when editing takes place during or after the embryo begins replicating its own DNA just before the first cell division. To help avoid mosaicism in this research, the guide, donor and Cas9 protein editing regents were introduced into bovine embryos just six hours after insemination, and before the beginning of DNA synthesis. One of the challenges with this timing for researchers hoping to insert a gene into the embryo is evading the cell’s predominant DNA repair pathway. In the early stages of embryo development, repair tends to be via the NHEJ pathway, whereas HDR editing efficiency is very low in early embryos.
The world’s first knock-in bull by CRISPR-mediated HMEJ in early embryos weighed 110 pounds (50kg) at birth.
An insertion strategy
To avoid the insertion of the new gene being thwarted by the cell’s DNA repair system, the team employed a homology-mediated end joining (HMEJ) strategy. In this process, HMEJ donor templates are designed so that the homology arms are flanked on both sides by the CRISPR target sequence. This HMEJ-based approach has proven to be effective in early stage 1-cell embryos, with a gene insertion efficiency much greater than that of alternative HDR-based strategies. To insert the SRY gene into the H11 locus, the team designed a HMEJ donor template, with the H11 CRISPR target sequence on either side of the homology arms.
However, insertion using this method is still not 100% efficient. In initial experiments, only around 40% of the bovine embryos were found to have the SRY insertion. Bovine embryo transfers, and the subsequent nine-month pregnancy, are expensive. This meant the team needed to confirm whether the gene insertion had been successful before the embryo was transferred to a surrogate cow. One way to do this is using embryonic biopsy, where a few cells are removed from the embryo and then checked for the presence of the inserted SRY gene. Unfortunately, this procedure can reduce the viability of the embryo, especially if the embryos are then frozen for transfer at a later date.
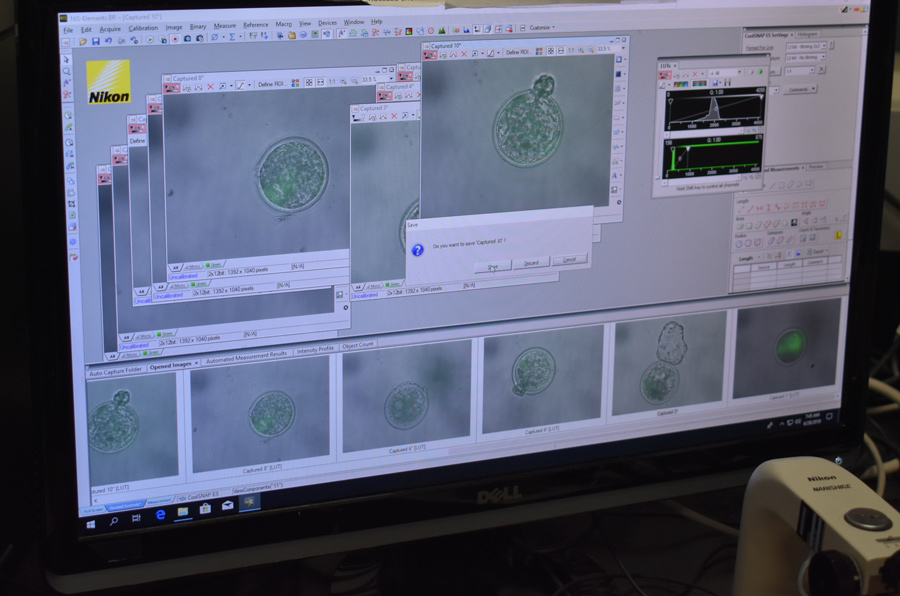
The team opted for a different screening approach: a “reporter” gene. A reporter gene carries an easily identifiable characteristic – fluorescence, for example – which allows for non-invasive screening of embryos. In this experiment, the jellyfish green fluorescent protein (GFP), was included alongside SRY, nestled between the homology arms on the HMEJ donor template. The fluorescent protein caused any embryo with an SRY insert to glow green when exposed to light in the blue to ultraviolet range. Embryos were screened, and in June 2019, nine 7-day embryos that fluoresced green under UV light were transferred to surrogate cows. A month later one of those cows, 3113, was confirmed pregnant, and an ultrasound scan revealed she was carrying a male bull calf.
Cosmo’s arrival
Nine months later, during the COVID-19 pandemic, Cosmo made his entrance. The world’s first knock-in bull by CRISPR-mediated HMEJ in early embryos weighed 110 pounds (50kg) at birth. The team were able to carry out detailed genetic analyses of the calf. They found that Cosmo carries several copies of the GFP:SRY gene on one of the Chromosome 17 pair, and a small insertion on the other. This suggests that, when Cosmo was a newly-formed embryo, the DSB introduced by Cas9 at the H11 locus on one Chromosome 17 was repaired by NHEJ resulting in a small 26 base pair insertion, and the other was repaired using the HMEJ-donor template to produce multiple GFP and SRY gene insertions, and a copy of the donor plasmid backbone.
The team hope that, one day in the future, Cosmo will become a father. Cosmo’s future offspring should reveal something interesting: will inheriting several copies of the SRY gene result in bovine XX embryos (which would normally be female) developing into male calves? And, of course, Cosmo still carries several copies of the green fluorescent reporter gene. However, in the absence of a giant darkened disco lounge where Cosmo could be exposed to light in the blue to ultraviolet range, the researchers will probably never be able to visualise Cosmo as a glowing “Green Bull”.
Personal Response
This is a fascinating story. What are the next steps for your research?
<>Fluorescent in situ hybridisation of chromosomal spreads will be used to confirm the chromosomal location and zygosity of the knock-in insertions. Hi-C, Illumina short read and PacBio long read data will be jointly used to investigate both the haplotype phase of Chromosome 17 and evaluate SNPs and alterations in the bull’s genome. We also have a couple more cows that are pregnant with calves for a project where genome editing was used to inactivate or knockout a gene.
This project highlights some of the complexities of working with large livestock species. Future analysis of the XX offspring inheriting the SRY gene on Chromosome 17 from this knock-in bull will reveal whether inheritance of the bovine SRY gene is sufficient to trigger the male developmental pathway in cattle. The 2-3 year generation interval of cattle means it will be years before we can characterise Cosmo’s offspring.
We are working on methods to try to improve the efficiency of genome editing by using an electric current, rather than microinjection, to introduce the editing reagents into bovine embryos. We hope this will increase the efficiency of the editing process, and improve the survivability of edited embryos.
Another complication is the current regulatory approach for gene edited livestock. In the United States, intentional genomic alterations introduced through gene editing in animals are regulated as veterinary drugs. This means they must go through a multigenerational safety and efficacy evaluation prior to entering commerce, which can become prohibitively expensive for small companies and academic researchers working with large livestock species.