X-ray ptychography: Seeing chromosome abnormalities in a new light
Chromosome abnormalities have profound health consequences including trisomy and cancer. Karyotyping (the visualisation of the complete set of chromosomes by shape and size) is an initial step towards identification of chromosome aberrations and disease. Other chromosome imaging methods such as fluorescent in situ hybridisation (FISH) rely on sample preparation or fluorescent markers, which may or may not alter the sample. Now Professor Ian Robinson of the London Centre for Nanotechnology (born of Imperial College London, Kings College London, and University College London) has investigated the masses of irradiated metaphase chromosomes using a novel imaging technique called X-ray ptychography, which can visualise unstained and unfixed samples, without the use of fluorescent probes.
In 1901, physics professor and mechanical engineer Wilhelm Conrad Roentgen won the first ever Nobel Prize in Physics. Six years earlier, Roentgen was conducting experiments with cathode rays (an electron beam in a vacuum tube which travels from a negatively charged electrode to a positively charged electrode) to determine whether they could pass through glass. As the cathode ray tube he was using was covered with thick black paper, he was surprised when a green light escaped, projected onto a fluorescent screen. He referred to this enigmatic green light as ‘X’ (or ‘unknown’) rays, and soon realised that they could pass through solid objects, including the soft tissues of the human body. One of the first X-rays (as they are now known) was an image of his wife’s hand, which clearly showed the bones of her hand and her wedding ring.
News of his discovery spread like wildfire around the world, and the phenomenon captured the imagination of the public. Roentgen did not patent the discovery, believing it to be too valuable to humankind. He had good reason – within a matter of years, doctors were using X-rays to detect broken bones, dental abysses, lung cancer, pneumonia, and more, and this paved the way for other more sophisticated forms of medical imaging such as computed tomography (CT), magnetic resonance imaging (MRI), echocardiograph, and ultrasound. In short, it became a cornerstone of modern medicine that we still use today. X-rays have moved on light years since this time, and scientists at University College London (UCL) are now using them to visualise tiny X-shaped structures that contain deoxyribonucleic acid (DNA) inside the nucleus of eukaryotic cells.

DNA: safeguarding our genetic information
DNA is the blueprint that carries the genetic code for most complex forms of life on this planet. Ribonucleic acid (RNA) is used by certain viruses, but is neither as stable nor as easily repaired as DNA. Both contain a form of sugar known as ‘ribose’ which consist of carbon atoms encircled by oxygen and hydrogen. DNA however, contains a ribose that has lost one oxygen and one hydrogen atom. This, along with the protective double helical structure of DNA, makes DNA an effective way to safeguard genetic information – the extra hydrogen and oxygen atoms present in RNA leaves it vulnerable to hydrolysis, a chemical reaction in which water breaks down complex molecules into simple molecules. As a result, RNA degrades approximately 100 times quicker than DNA. Illustrating this fact, scientists in 2021 discovered intact DNA fragments in the molars of Siberian steppe mammoths (a descendant of the woolly mammoth) that dates back to 1.2 million years ago. Prior to this discovery, the oldest genome ever sequenced was from a horse that died approximately 700,000 years ago.
The amount of DNA in each chromosome is known, but exactly which proteins make up the metaphase chromosome remains somewhat enigmatic.
The DNA found in chromosomes is tightly coiled around structural proteins called histones. Each healthy human cell contains 23 pairs of chromosomes (totalling 46) – one set from the mother and one set from the father. Chromosomes are only visible when the cell is undergoing mitosis (division), as this is when the DNA becomes more tightly packed.
Mitosis and the cell cycle
Mitosis is the process where cells undergo division and the DNA contained in the cell’s nucleus is divided and results in two new genetically identical daughter cells. There are four main sequential phases of mitosis: prophase (including prometaphase), metaphase, anaphase, and telophase. Cytokinesis (the physical division of the contents of the cell to create the daughter cells) occurs in the last two stages of the cycle. The stage just before mitosis is known as ‘interphase’ and this contains three main stages: G1 phase (where the cell grows larger, and copies its components), S phase (where the cell creates a copy of all the DNA in the nucleus of the cell), and G2 phase (where the cell produces organelles and prepares for mitosis).
Karyotype analysis: a diagnostic tool
Scientists can visualise a complete set of metaphase chromosomes in a particular organism or species – this is called the karyotype, in which the chromosomes are organised according to their size and shape. Analysis of the karyotype can reveal genetic abnormalities such as missing chromosomes, extra chromosomes, or genetic mutations such as deletions (where part of a chromosome or DNA sequence is omitted during DNA replication), duplications (another copy of the sequence is made), and translocations (a piece of a chromosome breaks and is re-attached to another chromosome). The medical implications of aberrant chromosomes are profound for the patient, with trisomy 21 (Down’s syndrome), trisomy 18 (Edward’s syndrome) and trisomy 13 (Patau syndrome) caused by extra chromosomal copies.

The amount of DNA in each chromosome is known, but exactly which proteins make up the metaphase chromosome remains somewhat enigmatic. As such, access to this knowledge would have important implications for diagnosing diseases with a genetic component, such as many human cancers.
Professor Ian Robinson led a group in the Research Complex at Harwell, which helped develop phase modulation technology for X-ray imaging, and built a beamline at National Synchrotron Light Source to develop the surface X-ray Diffraction technique. Robinson was awarded the Surface Structure Prize, the ACA Warren Prize, and the Gregori Aminoff Prize for his discovery of Crystal Truncation Rods, and the development of Bragg Coherent Diffractive Imaging.
Further to this research, Robinson has conducted a study at the London Centre for Nanotechnology (LCN) investigating the masses of each metaphase chromosome by measuring the phase shift of X-rays passing through them. The masses of the chromosomes were then measured after low-dose irradiation while the cells were still alive.

Ionising irradiation (energy that acts on living tissues by removing electrons from atoms and making them unstable) causes several different types of DNA damage which can lead to a wide range of diseases such as cancer. This DNA damage includes damage to chromosomes, which are known as numerical or structural abnormalities. A numerical abnormality means that a person would be missing one chromosome or has an extra copy of a chromosome. A structural abnormality, on the other hand, concerns a defect in the physical make up of that particular chromosome. These were the types of abnormalities investigated by Robinson using a cutting-edge imaging technique known as ‘X-ray ptychography’, in 2021.
X-ray ptychography: a novel chromosomal imaging method
In 2007, the ‘Diamond Light Source’ (DLS) facility was unveiled in Harwell, Oxfordshire. The research at this facility focused on using highly coherent X-rays to image biological matter and had a wide range of life-science applications. The same year, Professor John Rodenburg and colleagues invented the ‘X-ray ptychography’ method to amalgamate diffraction patterns obtained by a scanner beam into a useful, high-resolution image of a sample.
Ptychography is a method of microscopic imaging that uses a computer to generate a phase contrast image of a specimen. Ptychography can be used with visible light, X-ray, electrons, and extreme ultraviolet light. X-ray ptychography uses photons (tiny particles of light that carry waves of electromagnetic radiation) which interact with the inner shell of the atom under examination through refraction or absorption. Because it is based on optical interference, the method is able to map out the phase shifts due to interaction with the specimen.
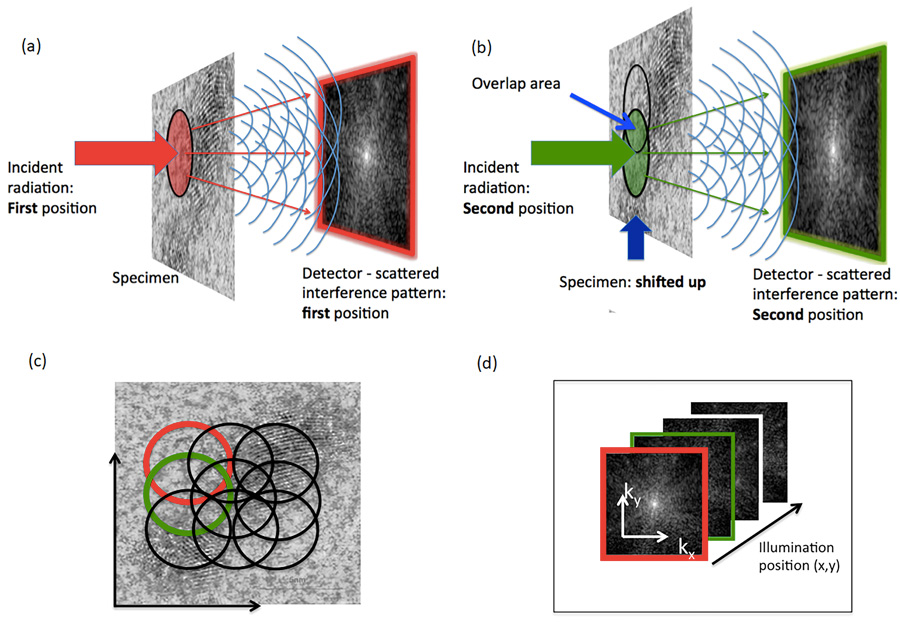
LCN researchers Archana Bhartiya and Yusuf Mohammed, developed a preparation method to protect biological samples from radiation damage. The chromosomes are then displayed in the form of ‘spreads’ during metaphase and their individual masses are compared with the known DNA length of each chromosome.
In Robinson’s study, human T-lymphocytes (white blood cells) were irradiated and harvested at metaphase for study using X-ray ptychography and the masses of each chromosome were then calculated. Even before irradiation, the masses of the chromosomes were 20 times heavier than the amount of DNA scientists know to be inside them from the human genome project. Most of the mass is made up of proteins that dictate how the DNA is coiled up into its characteristic structure, but even given this, the mass was larger than expected – which could indicate the presence of some unidentified proteins.
This study illustrated the response of living T-lymphocyte chromosomes to low dose irradiation; enough to cause breaks in the DNA strands, but below the 1 Gray, or Gy, limit that kills most cells. At 0.1Gy, it was noted that the mass of the chromosomes increased – this could be due to the DNA repair enzymes drafted to repair the damage done to the irradiated DNA. However, at 0.5Gy, a decrease in the mass of the chromosome was observed, followed by an increase at 1Gy. While these enigmatic results require further investigation, this technique represents a revolutionary step forward in the world of microscopic imaging.
X-ray ptychography uses photons which interact with the inner shell of the atom under examination through refraction or absorption.
Advantages of X-ray ptychography
Ptychography has several crucial advantages over conventional microscopy. Firstly, the imaging does not rely on a conventional lens, and therefore is unaffected by numerical aperture (its ability to obtain light and resolve fine specimen detail). Fluorescent in situ hybridisation (FISH) is another technique used for visualising chromosome abnormalities that is commercially available and is applied widely in medical practice. This method, however, relies on the use of a probe attached to a fluorescent marker, and the effects of this fluorescence are temporary.
Due to the imaging nature of X-ray ptychography, this technique also does not call for the specimen to be stained, fixed, or labelled with a fluorescent marker to be visible – sample preparation which carries with it a risk of damaging the natural presentation of the specimen.

The team is currently working to increase the sensitivity and accuracy of their technique, probing deeper into the fundamental building blocks of the human genome. This cutting-edge new technique has many highly promising clinical applications, and just as the invention of X-ray imaging did, it may pave the way for even more sophisticated microscopic imaging techniques for use in research and disease diagnosis.
Personal Response
Which medical or diagnostic application of this research do you think is most promising?
The physical structure of chromosomes and the mechanisms of how they coil up, copy themselves, and release the genetic information inside themselves is very much an active field of research. Numerous condensed states of the DNA-nucleosome complex have been discovered and may be relevant at different stages of the cell cycle. X-ray imaging has the important possibility of attaining high image resolutions of chromosomes trapped in these critical states. Knowing the structure will greatly increase our understanding of the mechanisms relevant to life, meaning cures can be devised for patients with a vast range of genetic diseases, including cancer.